1. Input file and data format¶
1.1. BED file¶
BED (Browser Extensible Data) format is commonly used to describe blocks of genome. The BED format consists of one line per feature, each containing 3-12 columns of data. It is 0-based (meaning the first base of a chromosome is numbered 0). It is s left-open, right-closed. For example, the bed entry “chr1 10 15” contains the 11-th, 12-th, 13-th, 14-th and 15-th bases of chromosome-1.
- BED12 file
- The standard BED file which has 12 fields. Each row in this file describes a gene or an array of disconnected genomic regions. Details are described here
- BED3 file
- Only has the first three required fields (chrom, chromStart, chromEnd). Each row is used to represent a single genomic region where “score” and “strand” are not necessary.
- BED3+ file
- Has at least three columns (chrom, chromStart, chromEnd). It could have other columns, but these additional columns will be ignored.
- BED6 file
- Has the first six fields (chrom, chromStart, chromEnd, name, score, strand). Each row is used to represent a single genomic region and their associated scores, or in cases where “strand” information is essential.
- BED6+ file
- Has at least six columns (chrom, chromStart, chromEnd, name, score, stand). It could have other columns, but these additional columns will be ignored.
1.2. Proportion values¶
In bisulfite sequencing (RRBS or WGBS), the methylation level of a particular CpG or region can be represented by a “proportion” value. We define the proportion value as a pair of integers separated by comma (“,”) with the first integer (m, 0 <- m <- n) representing “number of methylated reads” and the second integer (n, n >- 0) representing “number of total reads”. for example:
0,10 1,27 2,159 #Three proportions values indicated 3 hypo-methylated loci
7,7 17,19 30,34 #Three proportions values indicated 3 hyper-methylated loci
1.3. Beta values¶
The Beta-value is a value between 0 and 1, which can be interpreted as the approximation of the percentage of methylation for a given CpG or locus. One can convert proportion value into beta value, but not vice versa. In the equation below, C is the “probe intensity” or “read count” of methylated allele, while U is the “probe intensity” or “read count” of unmethylated allele.
1.4. M values¶
The M-value is calculated as the log2 ratio of the probe intensities (or read counts) of methylated allele versus unmethylated allele. In the equation below, C is the “probe intensity” or “read count” of methylated allele, while U is the “probe intensity” or “read count” of unmethylated allele. w is the offset or pseudo count added to both denominator and numerator to avoid unexpected big changes and performing log transformation on zeros.
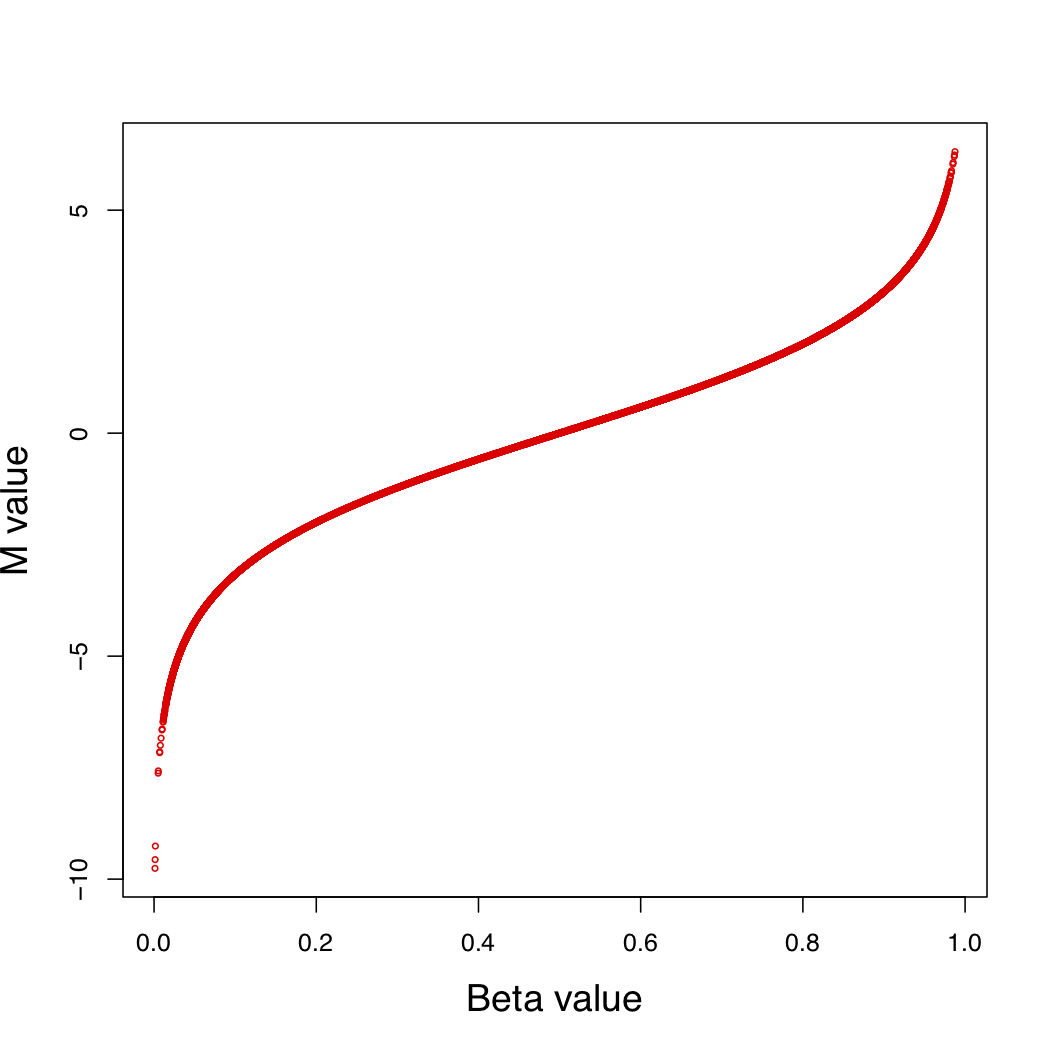
1.5. Convert Beta value to M value or vice versa¶
The relationship between Beta-value and M-value is shown as equation and figure: