15. beta_profile_region.py¶
15.1. Description¶
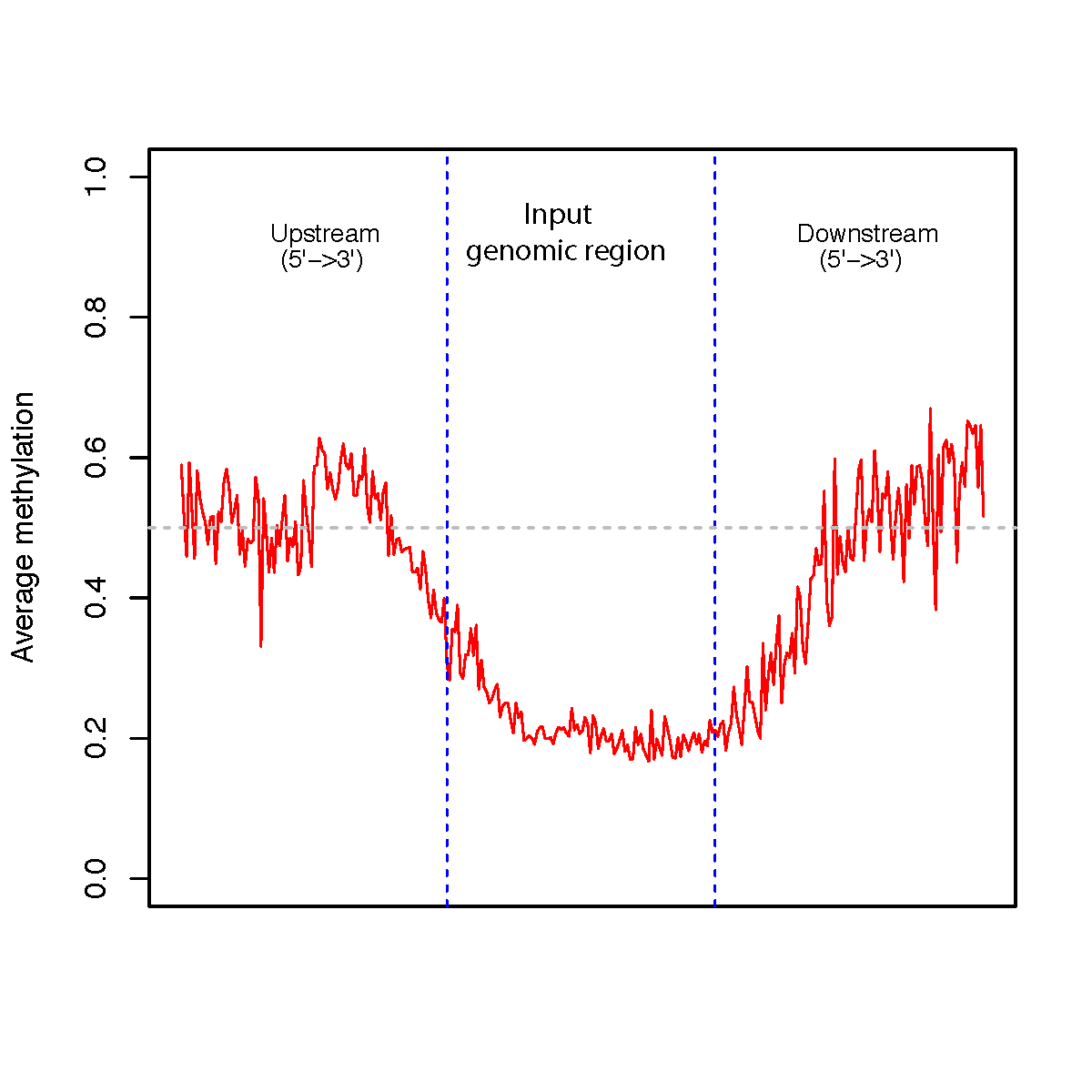
This program calculates methylation profile (i.e. average beta value) around the user-specified genomic regions.
Example of input
# BED6 format (INPUT_FILE)
chr22 44021512 44021513 cg24055475 0.9231 -
chr13 111568382 111568383 cg06540715 0.1071 +
chr20 44033594 44033595 cg21482942 0.6122 -
# BED3 format (REGION_FILE)
chr1 15864 15865
chr1 18826 18827
chr1 29406 29407
15.2. Options¶
--version show program’s version number and exit -h, --help show this help message and exit -i INPUT_FILE, --input_file=INPUT_FILE BED6+ file specifying the C position. This BED file should have at least six columns (Chrom, ChromStart, ChromeEnd, Name, Beta_value, Strand). BED6+ file can be a regular text file or compressed file (.gz, .bz2). -r REGION_FILE, --region=REGION_FILE BED3+ file of genomic regions. This BED file should have at least three columns (Chrom, ChromStart, ChromeEnd). If the 6-th column does not exist, all regions will be considered as on “+” strand. -d DOWNSTREAM_SIZE, --downstream=DOWNSTREAM_SIZE Size of extension to downstream. default=2000 (bp) -u UPSTREAM_SIZE, --upstream=UPSTREAM_SIZE Size of extension to upstream. default=2000 (bp) -o OUT_FILE, --output=OUT_FILE The prefix of the output file.
15.3. Input files (examples)¶
15.4. Command¶
$beta_profile_region.py -r hg19.RefSeq.union.1Kpromoter.bed.gz -i test_02.bed6.gz -o region_profile