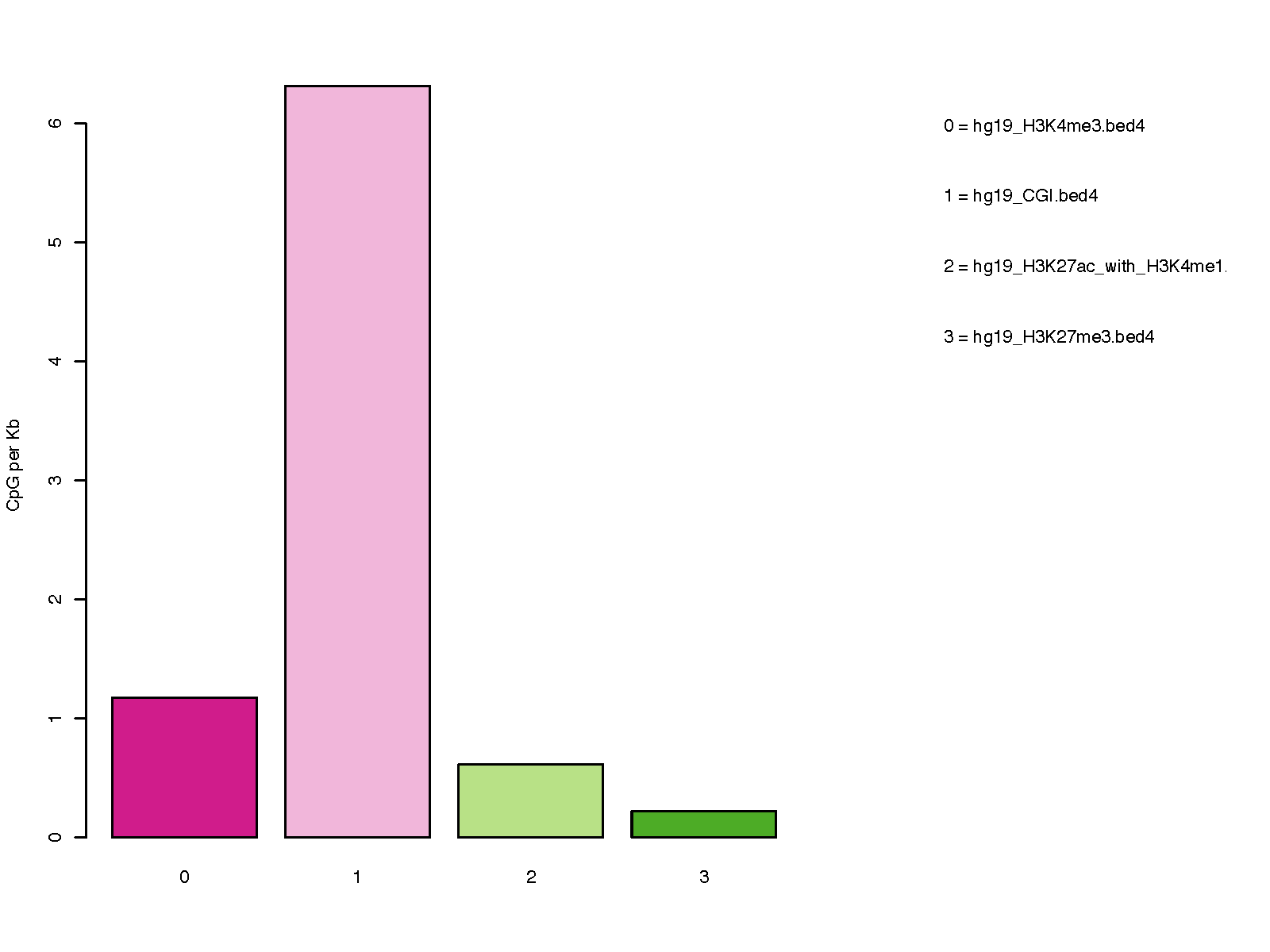
7. CpG_distrb_region.py¶
7.1. Description¶
This program calculates the distribution of CpG over user-specified genomic regions.
Notes
- A maximum of ten BED files (define ten different genomic regions) can be analyzed together.
- The order of BED files is important (i.e., considered as “priority order”). Overlapped genomic regions will be kept in the BED file with the highest priority and removed from BED files of lower priorities. For example, users provided 3 BED files via “-i promoters.bed,enhancers.bed,intergenic.bed”, then if an enhancer region is overlapped with promoters, the overlapped part will be removed from “enhancers.bed”.
- BED files can be regular or compressed by ‘gzip’ or ‘bz’.
7.2. Options¶
--version show program’s version number and exit -h, --help show this help message and exit -i CPG_FILE, --cpg=CPG_FILE BED file specifying the C position. This BED file should have at least three columns (Chrom, ChromStart, ChromeEnd). Note: the first base in a chromosome is numbered 0. This file can be a regular text file or compressed file (.gz, .bz2). -b BED_FILES, --bed=BED_FILES List of comma separated BED files specifying the genomic regions. -o OUT_FILE, --output=OUT_FILE The prefix of the output file.
7.3. Input files (examples)¶
- 850K_probe.hg19.bed3.gz Input bed file of 850K probe
- hg19_CGI.bed4 CpG islands
- hg19_H3K4me3.bed4 Promoters
- hg19_H3K27ac_with_H3K4me1.bed4 Bivalent promoters
- hg19_H3K27me3.bed4 Heterochromatin regions
7.4. Command¶
# check the distribution of 850K probes in 4 genomic regions (CpG islands, Promoters,
# Bivalent promoters, and Heterochromatin regions)
$CpG_distrb_region.py -i 850K_probe.hg19.bed3.gz -b hg19_H3K4me3.bed4,hg19_CGI.bed4,\
hg19_H3K27ac_with_H3K4me1.bed4,hg19_H3K27me3.bed4 -o regionDist