20. beta_trichotmize.py¶
20.1. Description¶
Rather than using a hard threshold to call “methylated” or “unmethylated” CpGs or regions, this program uses a probability approach (Bayesian Gaussian Mixture model) to trichotmize beta values into three status:
- Un-methylated : labeled as “0” in the result file
- Both the homologous chromosomes (i.e. The maternal and paternal chromosomes) are unmethylated.
- Semi-methylated : labeled as “1” in the result file
- Only one of the homologous chromosomes is methylated. This is also called allele-specific methylation or imprinting. Note: semi-methylation here is different from hemimethylation, which refers to “one of two (complementary) strands is methylated”.
- Full-methylated : labeled as “2” in the result file
- Both the homologous chromosomes (i.e., The maternal and paternal chromosomes) are methylated.
- unassigned : labeled as “-1” in the result file
- CpGs failed to assigned to the three categories above.
20.2. Algorithm¶
As described above, in somatic cells, most CpGs can be grouped into 3 categories including “Un-methylated”, “Semi-methylated (imprinted)” and “Full-methylated”. Therefore, the Beta distribution of CpGs can be considered as the mixture of 3 Gaussian distributions (i.e. components). beta_trichotmize.py first estimates the parameters (mu1, mu2, mu3) and (s1, s2, s3) of the 3 components using expectation–maximization (EM) algorithm, then it calculates the posterior probabilities ( p0, p1, and p2) of each component given the beta value of a CpG.
- p0
- the probability that the CpG belongs to un-methylated component.
- p1
- the probability that the CpG belongs to semi-methylated component.
- p2
- the probability that the CpG belongs to full-methylated component.
The classification will be made using rules:
if p0 == max(p0, p1, p2):
un-methylated
elif p2 == max(p0, p1, p2):
full-methylated
elif p1 == max(p0, p1, p2):
if p1 >= prob_cutoff:
semi-methylated
else:
unknown/unassigned
20.3. Options¶
--version show program’s version number and exit -h, --help show this help message and exit -i INPUT_FILE, --input_file=INPUT_FILE Input plain text file containing beta values with the 1st row containing sample IDs (must be unique) and the 1st column containing probe IDs (must be unique). -c PROB_CUTOFF, --prob-cut=PROB_CUTOFF Probability cutoff to assign a probe into “semi- methylated” class. default=0.99 -r, --report If True, generates “summary_report.txt” file. default=False -s RANDOM_STATE, --seed=RANDOM_STATE The seed used by the random number generator. default=99
20.4. Input files (examples)¶
20.5. Command¶
$beta_trichotmize.py -i test_05_TwoGroup.tsv -r
20.6. Output files¶
- .results.txt for each sample
- summary_report.txt
$ head CirrHCV_01.results.txt
#Prob_of_0: Probability of CpG belonging to un-methylation group
#Prob_of_1: Probability of CpG belonging to semi-methylation group
#Prob_of_2: Probability of CpG belonging to full-methylation group
#Assigned_lable: -1 = 'unassigned', 0 = 'un-methylation', 1 = 'semi-methylation', 2 = 'full-methylation'
Probe_ID Beta_value Prob_of_1 Prob_of_0 Prob_of_2 Assigned_lable
cg00000109 0.8776539440000001 0.05562534330044164 3.673659573888142e-93 0.9443746566995583 2
cg00000165 0.239308082 0.999222373166152 0.0007776268338481155 1.3380168478281785e-21 1
cg00000236 0.8951333909999999 0.052142920095512614 3.5462722261710256e-97 0.9478570799044873 2
cg00000292 0.783661275 0.22215555206863843 1.46921724055509e-72 0.7778444479313614 2
cg00000321 0.319783971 0.9999999909047641 9.09523558157906e-09 1.4703488768311725e-16 1
$ cat summary_report.txt
#means of components
Subject_ID Unmethyl SemiMethyl Methyl
CirrHCV_01 0.0705891104729628 0.4949428535816466 0.8694861885234295
CirrHCV_02 0.06775600800214297 0.5018649959502874 0.8731195740516192
CirrHCV_03 0.07063205540113326 0.49795240946021674 0.8730234341971185
...
#Weights of components
Subject_ID Unmethyl SemiMethyl Methyl
CirrHCV_01 0.27231055290074735 0.35186129618859385 0.3758281509106588
CirrHCV_02 0.2623073658620772 0.36736674559925425 0.37032588853866855
CirrHCV_03 0.2659211619015646 0.3563058727320757 0.37777296536635974
...
#Converge status and n_iter
Subject_ID Converged n_iter
CirrHCV_01 True 35
CirrHCV_02 True 37
CirrHCV_03 True 34
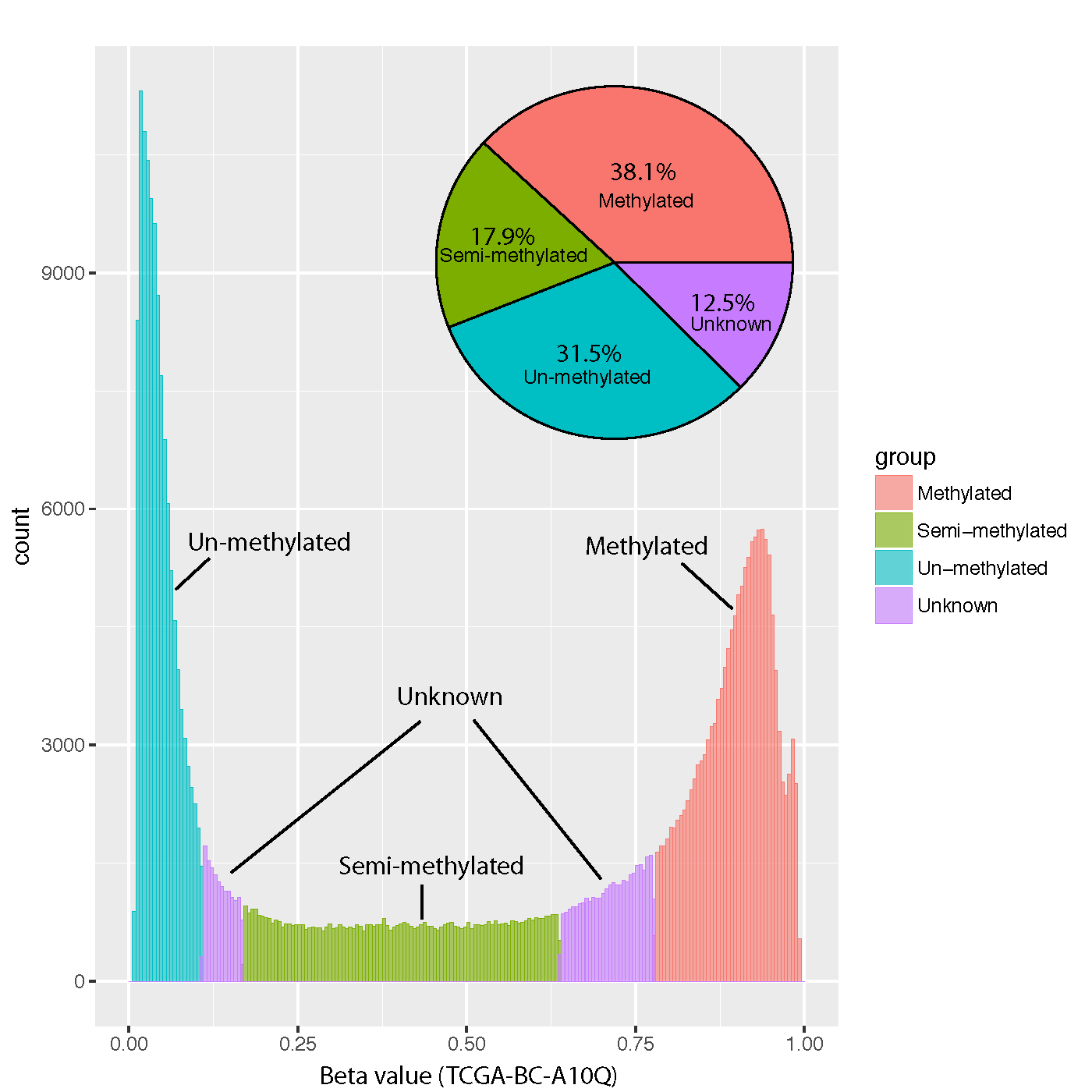
Below histogram and piechart showed the proportion of CpGs assigned to “Un-methylated”, “Semi-methylated” and “Full-methylated”.