10. beta_PCA.py¶
10.1. Description¶
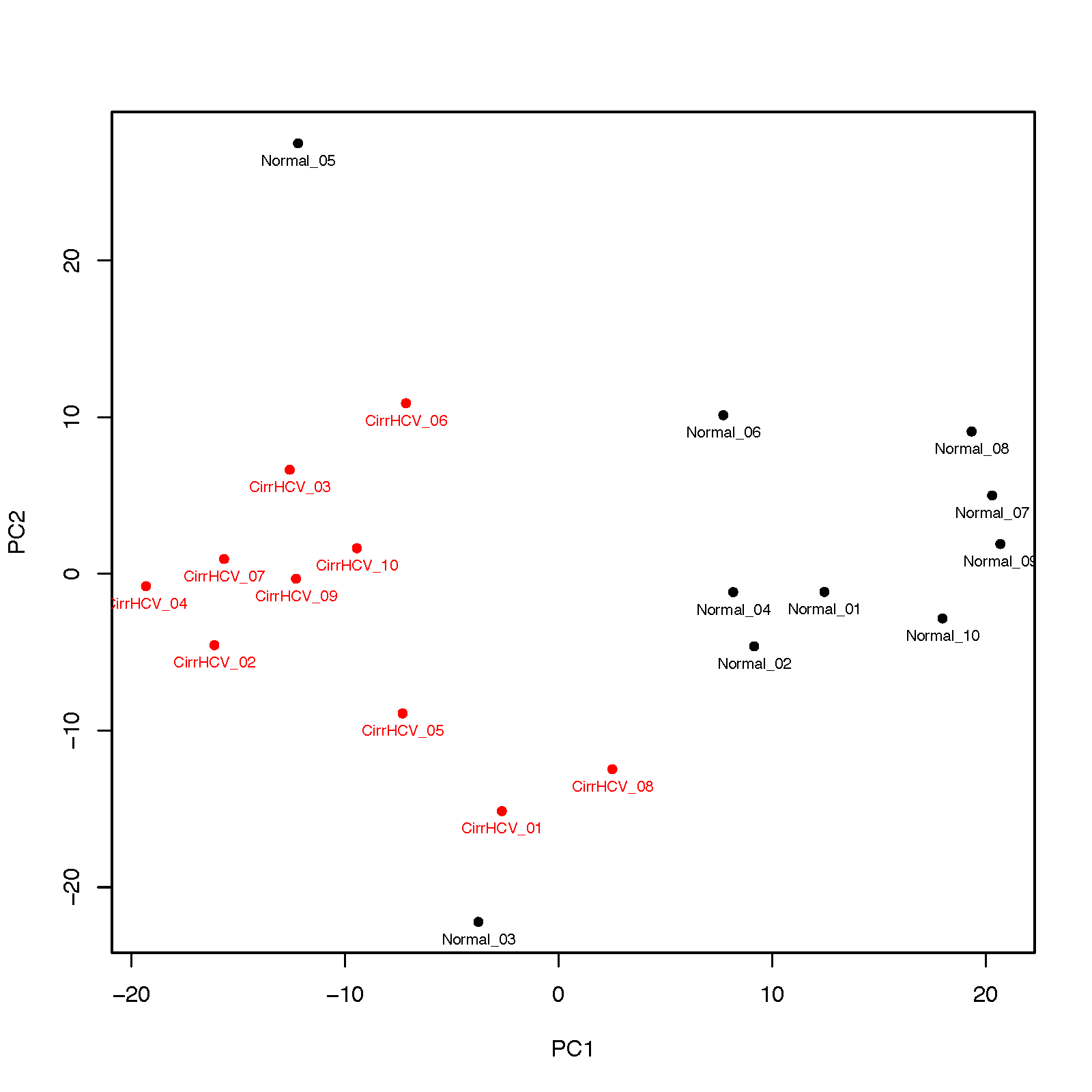
This program performs PCA (principal component analysis) for samples.
Example of input data file
ID Sample_01 Sample_02 Sample_03 Sample_04
cg_001 0.831035 0.878022 0.794427 0.880911
cg_002 0.249544 0.209949 0.234294 0.236680
cg_003 0.845065 0.843957 0.840184 0.824286
...
Example of input group file
Sample,Group
Sample_01,normal
Sample_02,normal
Sample_03,tumor
Sample_04,tumo
...
Notes
- Rows with missing values will be removed
- Beta values will be standardized into z scores
- Only the first two components will be visualized
- Variance% explained by each component will be printed to screen
- Options:
--version show program’s version number and exit -h, --help show this help message and exit -i INPUT_FILE, --input_file=INPUT_FILE Tab-separated data frame file containing beta values with the 1st row containing sample IDs and the 1st column containing CpG IDs. -g GROUP_FILE, --group=GROUP_FILE Comma-separated group file defining the biological groups of each sample. Different groups will be colored differently in the PCA plot. Supports a maximum of 20 groups. -n N_COMPONENTS, --ncomponent=N_COMPONENTS Number of components. default=2 -l, --label If True, sample ids will be added underneath the data point. default=False -c PLOT_CHAR, --char=PLOT_CHAR Ploting character: 1 = ‘dot’, 2 = ‘circle’. default=1 -a PLOT_ALPHA, --alpha=PLOT_ALPHA Opacity of dots. default=0.5 -x LEGEND_LOCATION, --loc=LEGEND_LOCATION Location of legend panel: 1 = ‘topright’, 2 = ‘bottomright’, 3 = ‘bottomleft’, 4 = ‘topleft’. default=1 -o OUT_FILE, --output=OUT_FILE The prefix of the output file.
10.2. Input files (examples)¶
10.3. Command¶
$beta_PCA.py -i cirrHCV_vs_normal.data.tsv -g cirrHCV_vs_normal.grp.csv -o HCV_vs_normal