5. CpG_distrb_chrom.py¶
5.1. Description¶
This program calculates the distribution of CpG over chromosomes
5.2. Options¶
--version show program’s version number and exit -h, --help show this help message and exit -i INPUT_FILES, --input_files=INPUT_FILES Input CpG file(s) in BED3+ format. Multiple BED files should be separated by “,” (eg: “-i file_1.bed,file_2.bed,file_3.bed”). BED file can be a regular text file or compressed file (.gz, .bz2). The barplot figures will NOT be generated if you provide more than 12 samples (bed files). [required] -n FILE_NAMES, --names=FILE_NAMES Shorter and meaningful names to label samples. Should be separated by “,” and match CpG BED files in number. If not provided, basenames of CpG BED files will be used to label samples. [optional] -s CHROM_SIZE, --chrom-size=CHROM_SIZE Chromosome size file. Tab or space separated text file with two columns: the first column is chromosome name/ID, the second column is chromosome size. This file will determine: (1) which chromosomes are included in the final bar plots, so do NOT include ‘unplaced’, ‘alternative’ contigs in this file. (2) The order of chromosomes in the final bar plots. [required] -o OUT_FILE, --output=OUT_FILE The prefix of the output file. [required]
5.3. Input files (examples)¶
5.4. Command¶
$ chrom_distribution.py -i 450K_probe.hg19.bed3.gz,850K_probe.hg19.bed3.gz -n 450K,850K \
-s hg19.chrom.sizes -o chromDist
Output files
- chromDist.txt
- chromDist.r
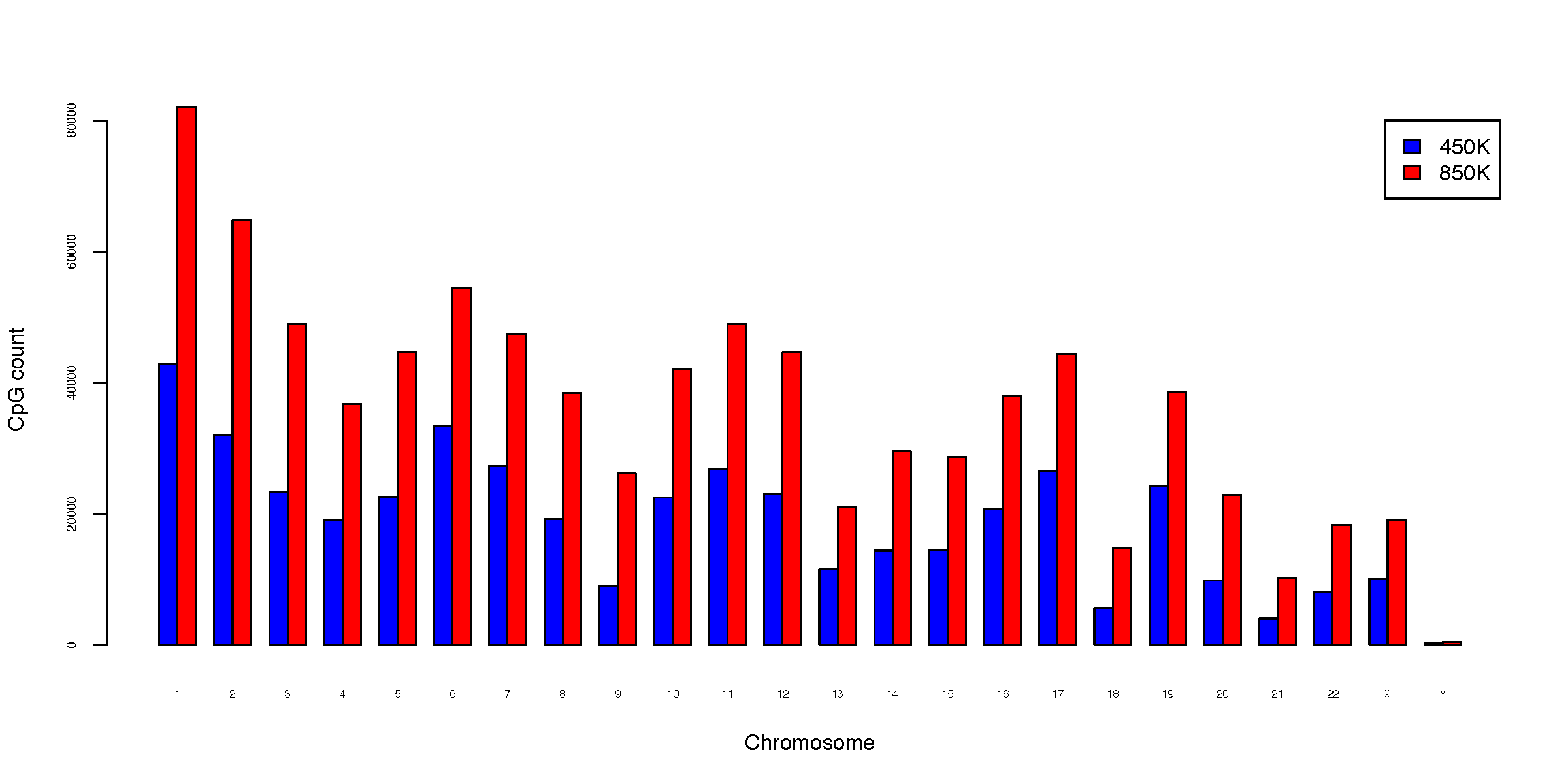
- chromDist.CpG_total.pdf
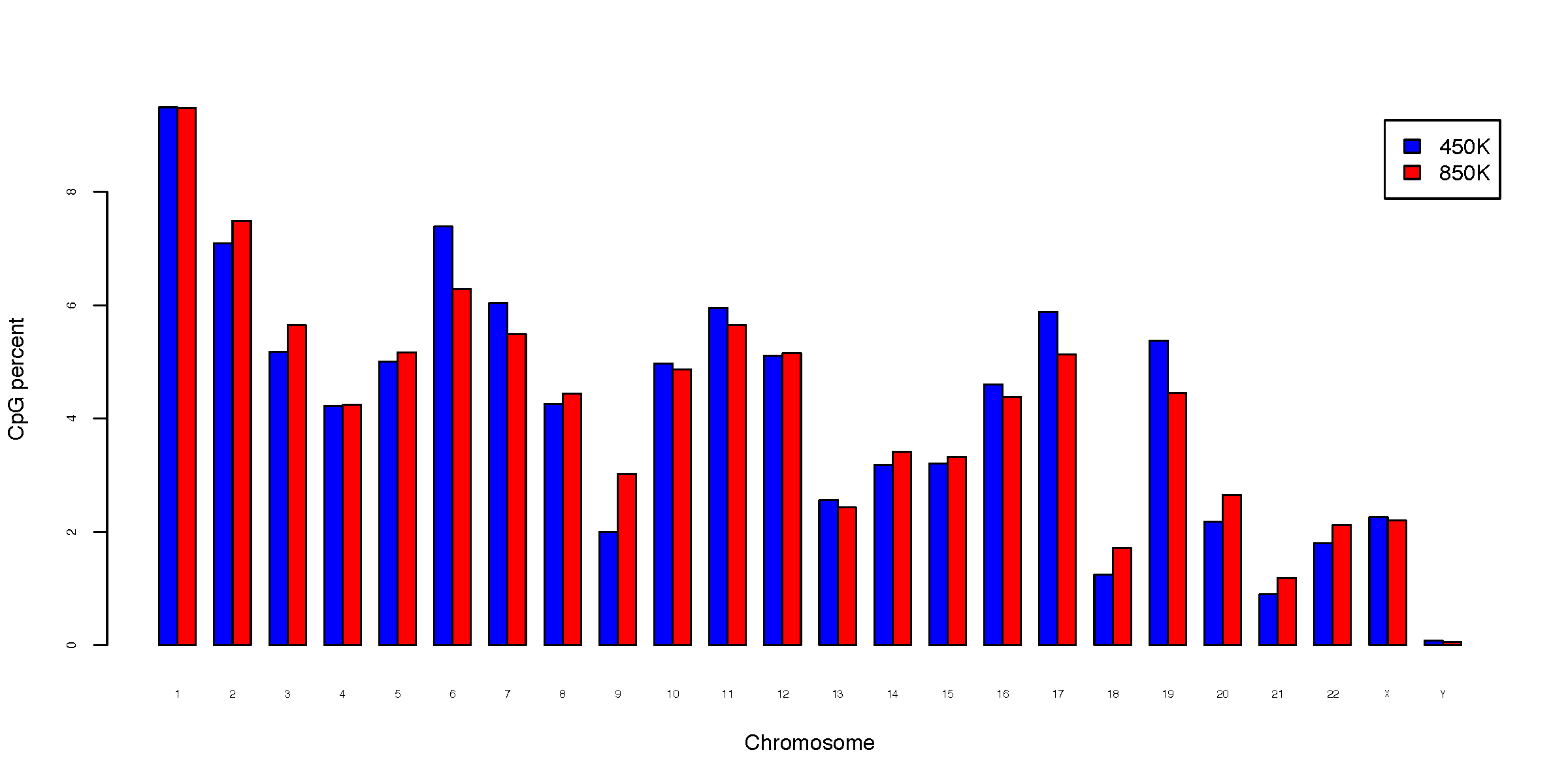
- chromDist.CpG_percent.pdf
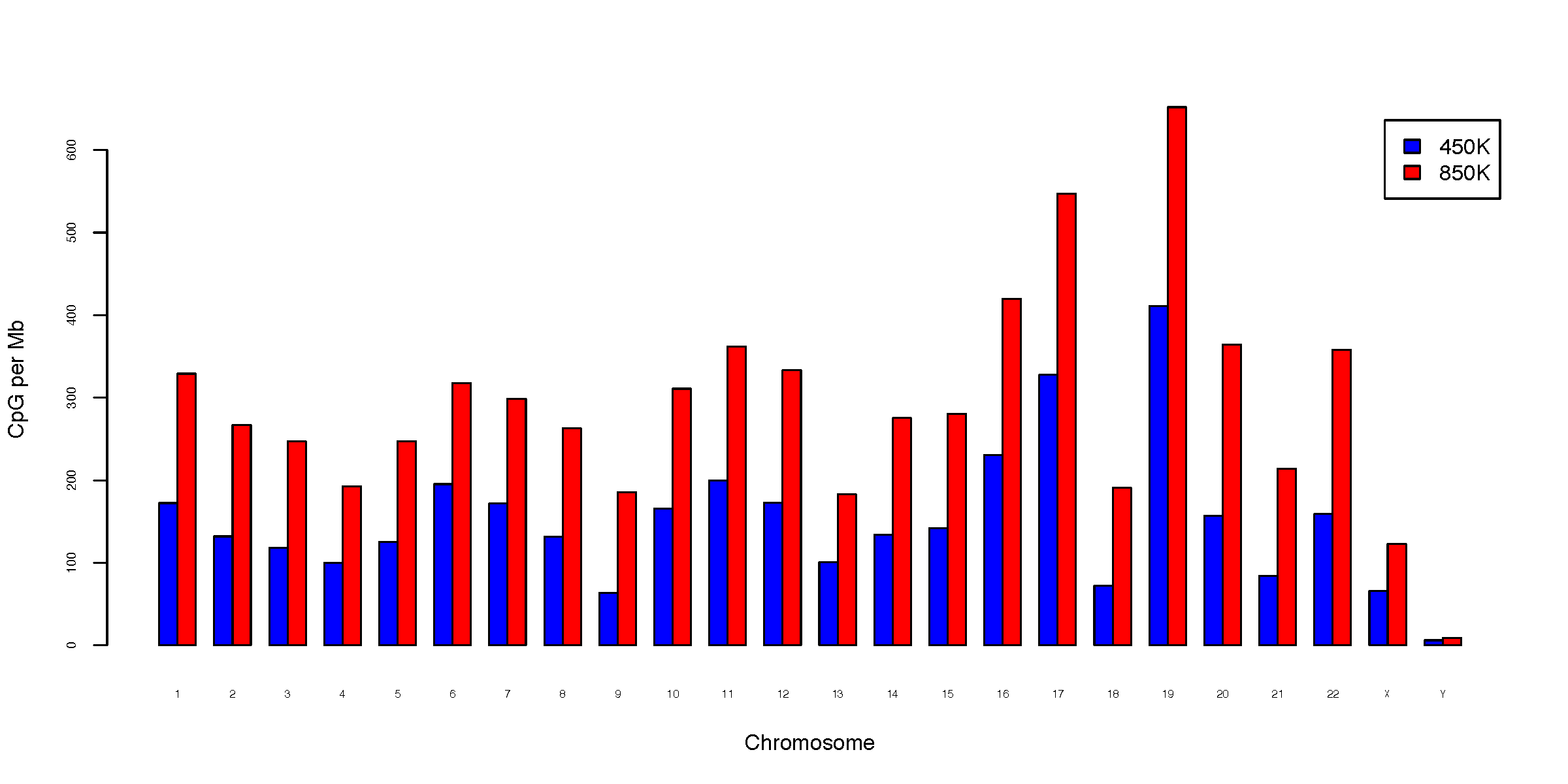
- chromDist.CpG_perMb.pdf
Total CpG count per chromosome
CpG percent on each chromosome (normalized to total CpGs)
CpG per Mb (normalized to chromosome size)