8. CpG_logo.py¶
8.1. Description¶
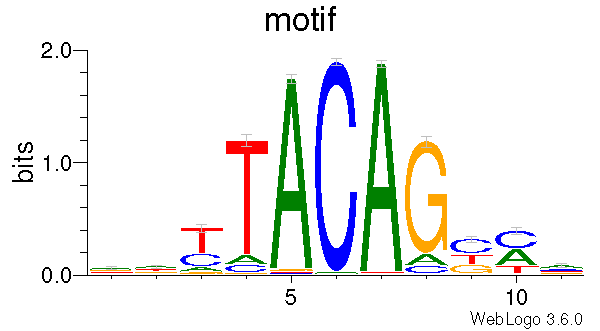
This program generates a DNA motif logo for a given set of CpGs. To answer the question of “what is the genomic context for a given list of CpGs ?”. This program first extracts genomic sequences around C position, and then generate motif matrices include:
- position frequency matrix (PFM)
- position probability matrix (PPM)
- position weight matrix (PWM)
- MEME format matrix
- Jaspar format matrix
It also generates motif logo using weblogo
Notes
- input BED file must have strand information.
8.2. Options¶
--version show program’s version number and exit -h, --help show this help message and exit -i INPUT_FILE, --input_file=INPUT_FILE BED file specifying the C position. This BED file should have at least six columns (Chrom, ChromStart, ChromeEnd, name, score, strand). Note: Must provide correct strand information. This file can be a regular text file or compressed file (.gz, .bz2). -r GENOME_FILE, --refgenome=GENOME_FILE Reference genome seqeunces in FASTA format. Must be indexed using the samtools “faidx” command. -e EXTEND_SIZE, --extend=EXTEND_SIZE Number of bases extended to up- and down-stream. default=5 (bp) -n MOTIF_NAME, --name=MOTIF_NAME Motif name. default=motif -o OUT_FILE, --output=OUT_FILE The prefix of the output file.
8.3. Input files (examples)¶
- Human reference genome sequences in FASTA format: hg19.fa.gz and hg38.fa.gz
- 450_CH.hg19.bed.gz
8.4. Command¶
$CpG_logo.py -i 450_CH.hg19.bed.gz -r hg19.fa -o 450_CH
8.5. Output files¶
- 450_CH.logo.fa
- 450_CH.logo.jaspar
- 450_CH.logo.meme
- 450_CH.logo.pfm
- 450_CH.logo.ppm
- 450_CH.logo.pwm
- 450_CH.logo.logo.pdf